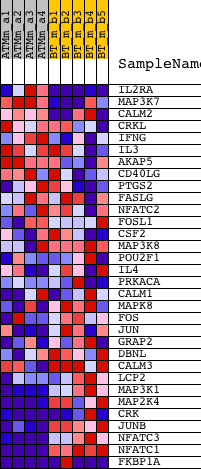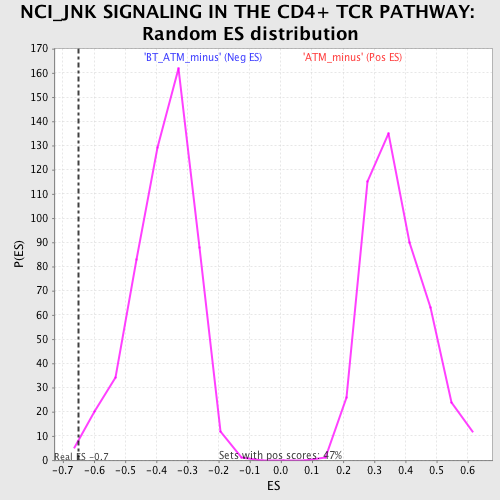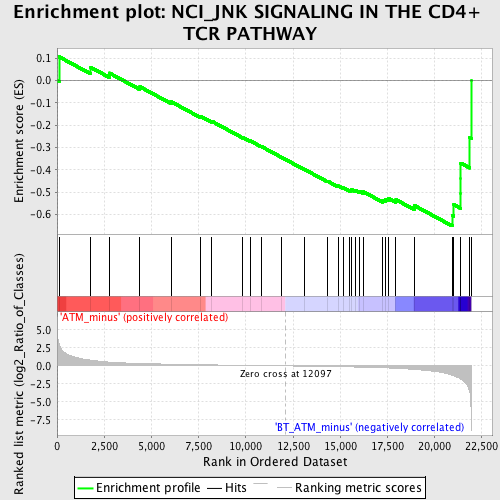
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | NCI_JNK SIGNALING IN THE CD4+ TCR PATHWAY |
| Enrichment Score (ES) | -0.65136117 |
| Normalized Enrichment Score (NES) | -1.7273109 |
| Nominal p-value | 0.0037453184 |
| FDR q-value | 0.16254598 |
| FWER p-Value | 0.686 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IL2RA | 1420691_at 1420692_at | 108 | 2.955 | 0.1054 | No | ||
| 2 | MAP3K7 | 1419988_at 1425795_a_at 1426627_at 1449693_at 1455441_at 1458787_at | 1785 | 0.784 | 0.0581 | No | ||
| 3 | CALM2 | 1422414_a_at 1423807_a_at | 2762 | 0.525 | 0.0332 | No | ||
| 4 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 4385 | 0.333 | -0.0285 | No | ||
| 5 | IFNG | 1425947_at | 6045 | 0.227 | -0.0957 | No | ||
| 6 | IL3 | 1450566_at | 7599 | 0.155 | -0.1608 | No | ||
| 7 | AKAP5 | 1447301_at | 8177 | 0.134 | -0.1822 | No | ||
| 8 | CD40LG | 1422283_at | 9841 | 0.076 | -0.2553 | No | ||
| 9 | PTGS2 | 1417262_at 1417263_at | 10217 | 0.063 | -0.2700 | No | ||
| 10 | FASLG | 1418803_a_at 1449235_at | 10804 | 0.043 | -0.2952 | No | ||
| 11 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 11899 | 0.006 | -0.3449 | No | ||
| 12 | FOSL1 | 1417487_at 1417488_at | 13122 | -0.038 | -0.3992 | No | ||
| 13 | CSF2 | 1427429_at | 14310 | -0.086 | -0.4502 | No | ||
| 14 | MAP3K8 | 1419208_at | 14885 | -0.112 | -0.4722 | No | ||
| 15 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 15180 | -0.126 | -0.4810 | No | ||
| 16 | IL4 | 1449864_at | 15504 | -0.141 | -0.4904 | No | ||
| 17 | PRKACA | 1447720_x_at 1450519_a_at | 15605 | -0.147 | -0.4895 | No | ||
| 18 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 15802 | -0.158 | -0.4926 | No | ||
| 19 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 16000 | -0.170 | -0.4952 | No | ||
| 20 | FOS | 1423100_at | 16233 | -0.185 | -0.4989 | No | ||
| 21 | JUN | 1417409_at 1448694_at | 17249 | -0.260 | -0.5356 | No | ||
| 22 | GRAP2 | 1456432_at | 17380 | -0.272 | -0.5314 | No | ||
| 23 | DBNL | 1460334_at | 17553 | -0.289 | -0.5284 | No | ||
| 24 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 17944 | -0.333 | -0.5338 | No | ||
| 25 | LCP2 | 1418641_at 1418642_at | 18938 | -0.496 | -0.5606 | No | ||
| 26 | MAP3K1 | 1424850_at | 20926 | -1.284 | -0.6034 | Yes | ||
| 27 | MAP2K4 | 1426233_at 1442679_at 1451982_at | 21011 | -1.366 | -0.5563 | Yes | ||
| 28 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 21375 | -1.800 | -0.5056 | Yes | ||
| 29 | JUNB | 1415899_at | 21378 | -1.811 | -0.4381 | Yes | ||
| 30 | NFATC3 | 1419976_s_at 1452497_a_at | 21380 | -1.817 | -0.3703 | Yes | ||
| 31 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 21857 | -3.609 | -0.2573 | Yes | ||
| 32 | FKBP1A | 1416036_at 1438795_x_at 1438958_x_at 1448184_at 1456196_x_at | 21931 | -6.986 | 0.0001 | Yes |